Thesis Plots
thesis_plots.RmdGlobal settings:
- theme
- figure width (= latex textwidth)
textwidth <- 6.14
# wes_palette("Zissou1")2 Background
Relative sizes of things on a logarithmic scale:
# define locations of vertical dashed lines
vLines <- c("atom" = 0.1e-9, "DNA" = 2e-9, "protein" = 4.2e-9, "phage" = 55e-9, "virus" = 100e-9, "bacteria" = 3e-6, "animal_cell" = 20e-6, "plant_cell" = 80e-6, "drosophilia" = 3e-3, "human" = 1.8)
vLines
#> atom DNA protein phage virus bacteria
#> 1.0e-10 2.0e-09 4.2e-09 5.5e-08 1.0e-07 3.0e-06
#> animal_cell plant_cell drosophilia human
#> 2.0e-05 8.0e-05 3.0e-03 1.8e+00
# define locations of range dashed lines
rangesXmin <- c(3e-9, 10e-6, 10e-6, 10e-6)
rangesXmax <- c(6e-9, 30e-6, 100e-6, 1e-3)
rangesY <- c(0.05, 0.1, 0.2, 0.13)
# x coordinates for axis labels
xBreaks <- 10^seq(-10, 1)
names(xBreaks) <- c("0.1 nm", "1 nm", "10 nm", "100 nm", "1 µm", "10 µm", "100 µm", "1 mm", "10 mm", "100 mm", "1 m")
ggplot() +
geom_vline(xintercept = vLines, alpha = 0.2, linetype = "dashed") +
geom_errorbarh(aes(xmin = rangesXmin, xmax = rangesXmax, y = rangesY), alpha = 0.2, linetype = "dashed", height = 0.05) +
# geom_rect(aes(xmin = 10e-6, xmax = 30e-6, ymin = 0, ymax = Inf), alpha = 0.2) +
# geom_tile(aes(x = 20e-6, y = 0, width = 20e-6, height = Inf), color = "red") +
scale_x_log10(breaks = xBreaks) +
scale_y_continuous(breaks = c(), limits = c(0,1), expand = expansion(add = 0)) +
theme_pretty_thesis() +
theme(axis.title.y = element_blank())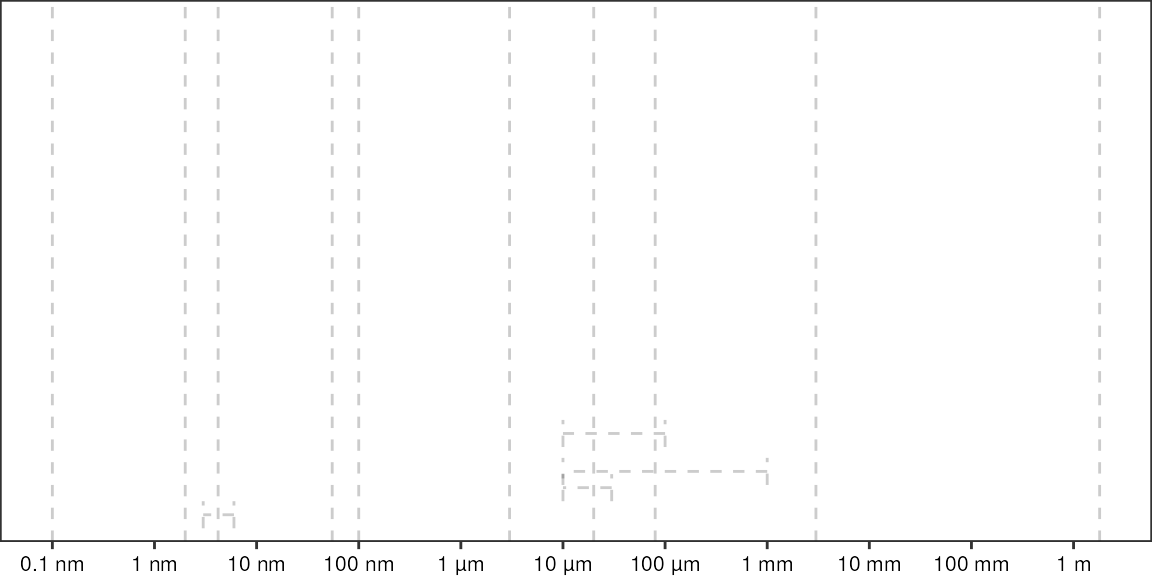
# annotation_logticks(side = "b", color = "gray")Save as PDF:
grDevices::pdf(file = "output/plots/02_relative_sizes.pdf", width = textwidth, height = textwidth/3, )
print(last_plot())
grDevices::dev.off()
#> agg_png
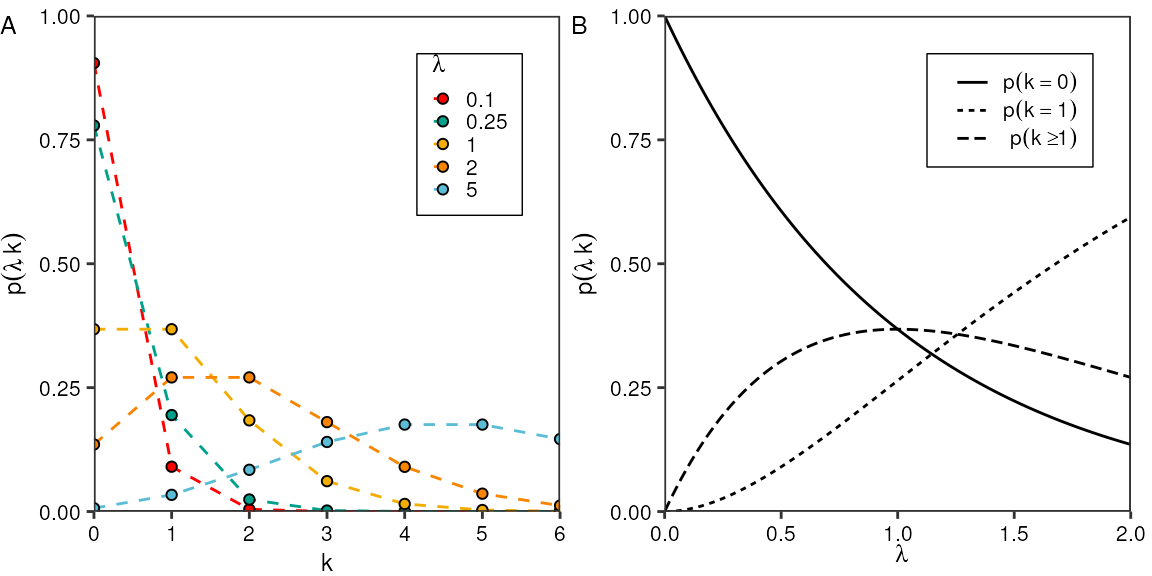
#> 2Poisson distribution for different \(\lambda\):
p1 <- tidyr::crossing("k" = seq(0,6),
"lambda" = c(0.1, 0.25, 1, 2, 5)) %>%
dplyr::mutate("p" = dpois(k, lambda)) %>%
ggplot(aes(x = k, y = p)) +
geom_line(aes(color = factor(lambda)), linetype = "dashed") +
geom_point(aes(fill = factor(lambda)), shape = 21) +
scale_x_continuous(breaks = seq(0,6)) +
# scale_y_continuous(breaks = c(0, 0.2, 0.4, 0.6, 0.8, 1)) +
theme_pretty_thesis() +
scale_fill_manual(values = wes_palette("Darjeeling1")) +
scale_color_manual(values = wes_palette("Darjeeling1")) +
theme(
# panel.grid.major = element_line(color = "black", linewidth = 0.1),
legend.position = c(1,1),
legend.justification = c(1,1)) +
labs(y = expression(p(lambda, k)),
x = "k",
color = expression(lambda),
fill = expression(lambda),
tag = "A") +
ylim(c(0,1)) +
coord_cartesian(expand =F)
p1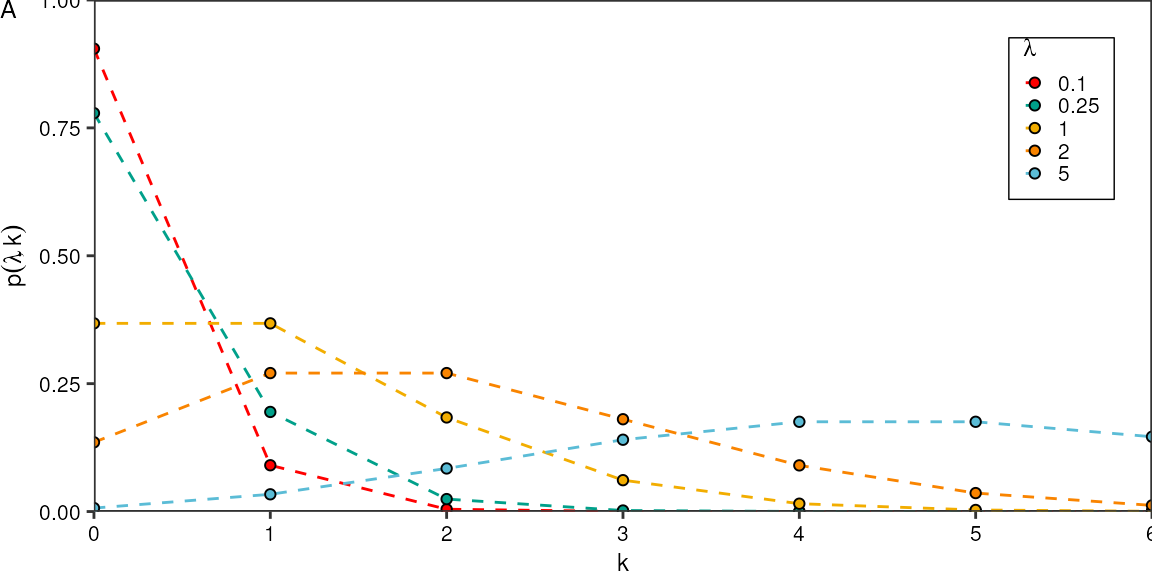
Evaluate Poisson PDF as function of lambda:
p2 <- tibble::tibble("lambda" = seq(0,2,0.01),
"single" = stats::dpois(1, lambda),
"empty" = stats::dpois(0, lambda),
"multiple" = 1 - stats::ppois(1, lambda)) %>%
tidyr::pivot_longer(!lambda) %>%
ggplot(aes(x = lambda, y = value, linetype = name)) +
geom_line() +
scale_linetype_discrete(labels = c(expression(p(k == 0)), expression(p(k==1)), expression(p(k >= 1))),
guide = guide_legend(title = NULL, title.position = "left")) +
theme_pretty_thesis() +
theme(legend.position = c(1,1),
legend.justification = c(1,1),
legend.key.width = unit(0.5, "cm")) +
coord_cartesian(expand = F) +
labs(x = expression(lambda),
y = expression(p(lambda, k)),
tag = "B")
p2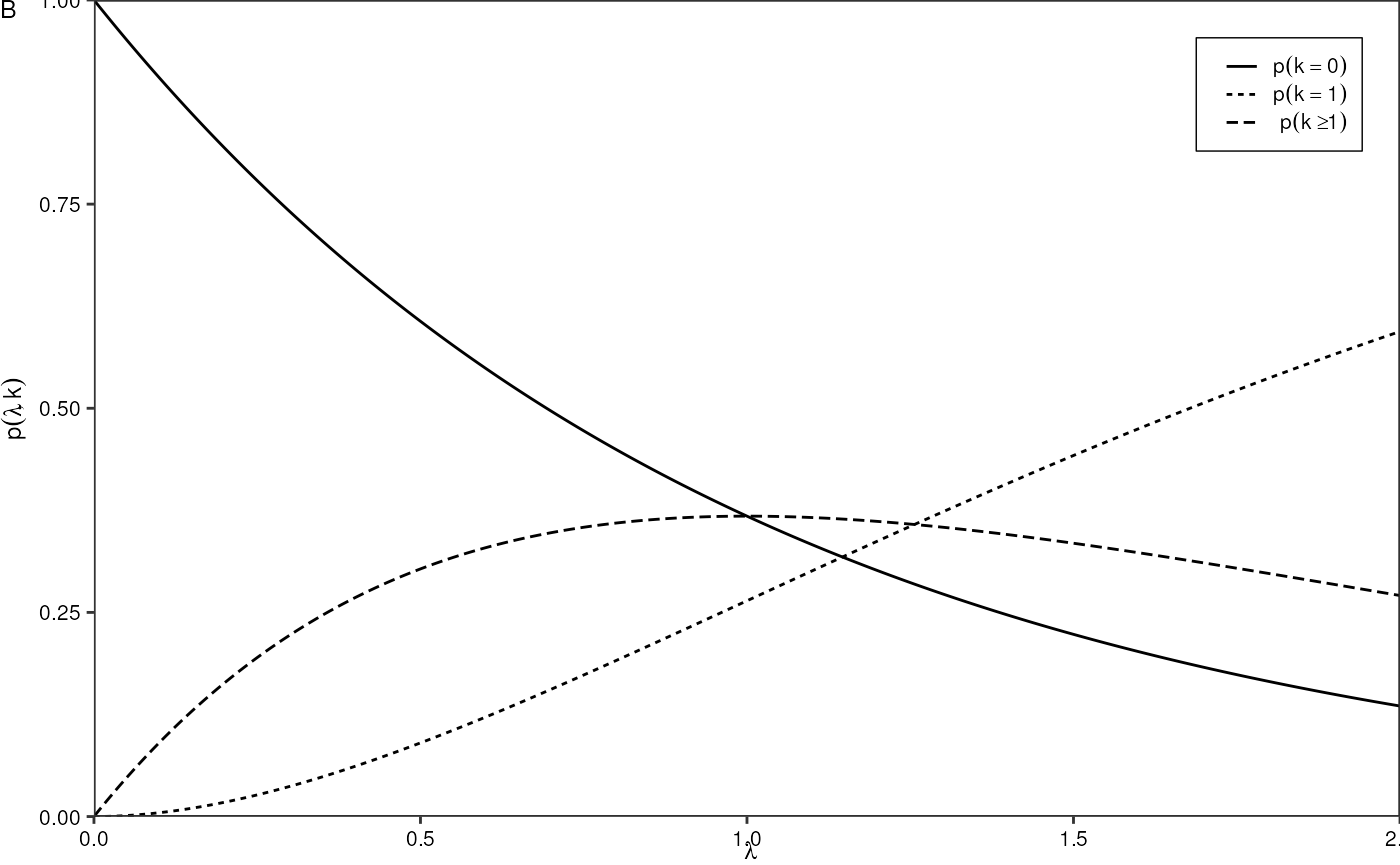
Patchwork:
4 Results
fads <- fads_read_tsv("data/fads/220916/pi30v4_6.txt")
fads
#> # A tibble: 413,790 × 6
#> time blue green red width spacing
#> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
#> 1 3647025511. 0 0.0961 0.723 0.32 0.915
#> 2 3647025516. 0 0.0958 0.763 0.37 0.665
#> 3 3647025520. 0 0.125 0.772 0.37 0.495
#> 4 3647025522. 0 0.357 0.759 0.355 0.025
#> 5 3647025526. 0 0.101 0.748 0.345 0.255
#> 6 3647025530. 0 0.0995 0.714 0.285 0.585
#> 7 3647025534. 0 0.0967 0.730 0.33 0.41
#> 8 3647025538. 0 0.0946 0.763 0.4 0.435
#> 9 3647025543. 0 0.0958 0.752 0.355 0.695
#> 10 3647025547. 0 0.0977 0.769 0.405 0.415
#> # … with 413,780 more rows
fads %>%
fads_remove_outliers() %>%
ggplot(aes(x = green, y = red)) +
geom_bin2d(bins = 100, drop = T) +
scale_fill_gradientn(colours = wes_palette("Zissou1", 100, type = "continuous")) +
theme_pretty_thesis() +
theme(legend.position = c(1, 0),
legend.justification = c(1,0),
legend.box.background = element_rect(fill = "white")) +
labs(x = "Green PMT signal [V]",
y = "Red PMT signal [V]",
fill = "Droplets")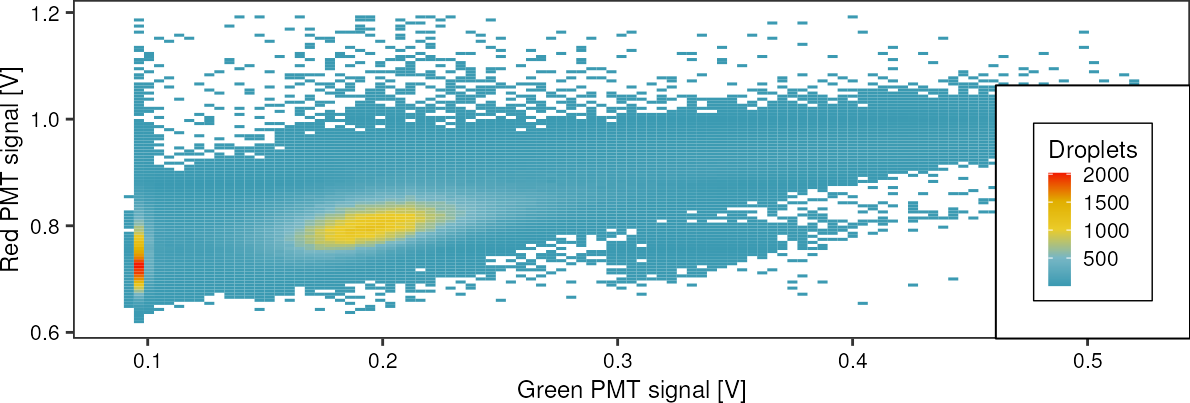
Supplementary Information
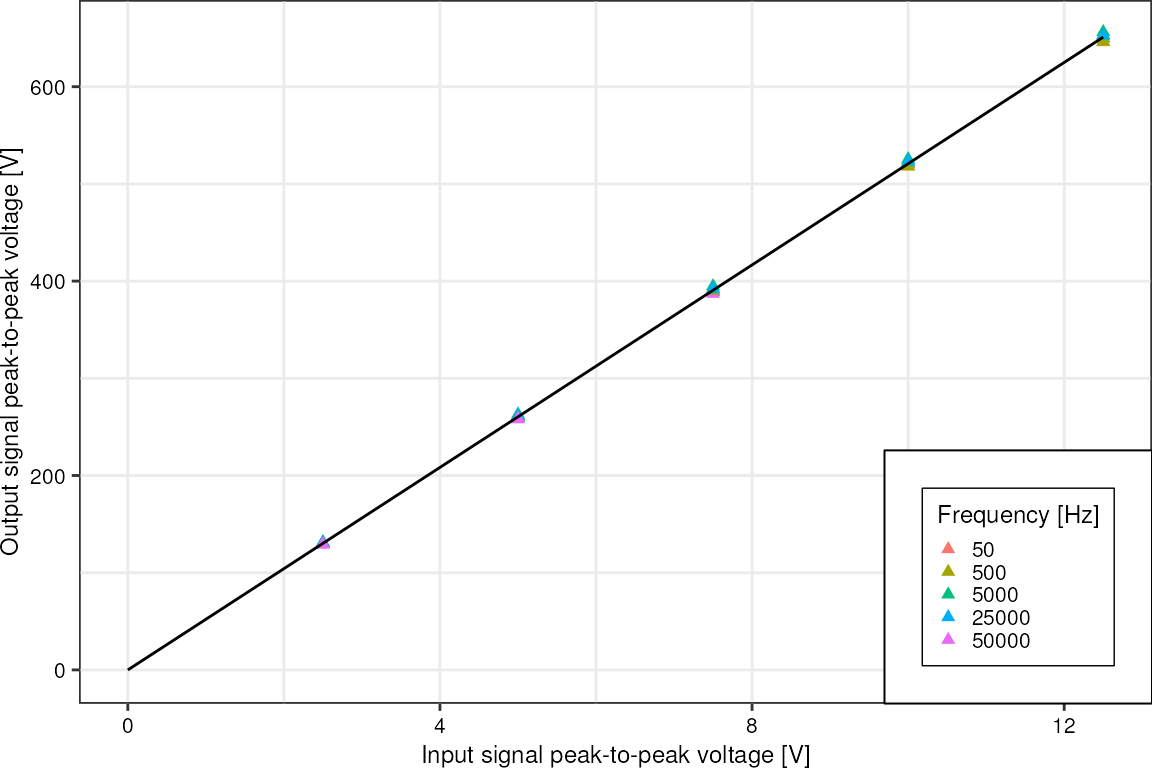
Voltage amplifier:
d <- readr::read_csv("data/boexle_sorter_output.csv", show_col_types = FALSE)
p <- ggplot(d, aes(x = input_vpp, y = output_vpp)) +
geom_point(aes(color = factor(frequency)), shape = 17) +
geom_smooth(method = "lm", formula = "y ~ x", fullrange = TRUE, color = "black", linewidth = 0.5, alpha = 0.3) +
xlim(c(0, 12.5)) +
labs(x = "Input signal peak-to-peak voltage [V]",
y = "Output signal peak-to-peak voltage [V]") +
theme_pretty_thesis() +
theme(panel.grid.major = element_line(),
panel.grid.minor = element_line(),
legend.box.background = element_rect(fill = "white")) +
labs(color = "Frequency [Hz]")
p